Since scientists first sequenced the human genome nearly 20 years ago, we have all been increasingly enthusiastic about the potential for precision medicine to better diagnose and treat diseases. In oncology, targeted therapies against specific mutations have already demonstrated this promise, though only in the relatively limited settings where these therapies exist and testing is done as part of the standard of care (like HER2 in breast or EGFR in lung).
Most cancer patients do not have the opportunity to get tested or to receive appropriate targeted therapies. So, accelerating pharmaceutical and diagnostic development has enormous potential to improve outcomes and save lives.
In this 3-part series, we further explore the challenges and potential of evolving and integrating precision medicine into oncology:
- Part I: The Science Behind Molecular Testing
- Part II: Current Clinical Utility and Commercial Challenges
- Part III: Future Co-Evolution of Diagnostics and Pharma
Key terms:
- Antibody: a type of protein which binds very selectively to another specific protein (used to detect mutated proteins and measure protein expression levels)
- DNA: linear strand of base pairs; the sequence of which encodes genetic information used to build proteins
- Diagnostic: a test for molecular alterations that indicates or confirms a disease or specific molecular alteration
- Gene: a stretch of DNA that encodes a protein
- Gene / Genomic (or DNA) Sequencing: process of reading and reporting out the DNA or mRNA sequence (to check for mutations)
- Molecular alteration: any change in a protein or its precursors that impedes its normal biological function. This includes both mutations that change the function of a protein, as well as significant increases or decreases in the amount of the protein.
- Molecular test: a test that examines DNA, mRNA, or protein to detect molecular alterations
- mRNA: a mobile copy of a gene that is transported throughout the cell to make proteins; the amounts of mRNA loosely corresponds to protein amounts
- Protein: a chain of amino acids (built based on a DNA/mRNA sequence) folded into a 3-dimensional molecule that works as a cellular machine to carry out biological processes
- Precision medicine: collecting information about molecular alterations and incorporating it into treatment decision-making
Overview of Molecular Alterations and Testing Technology
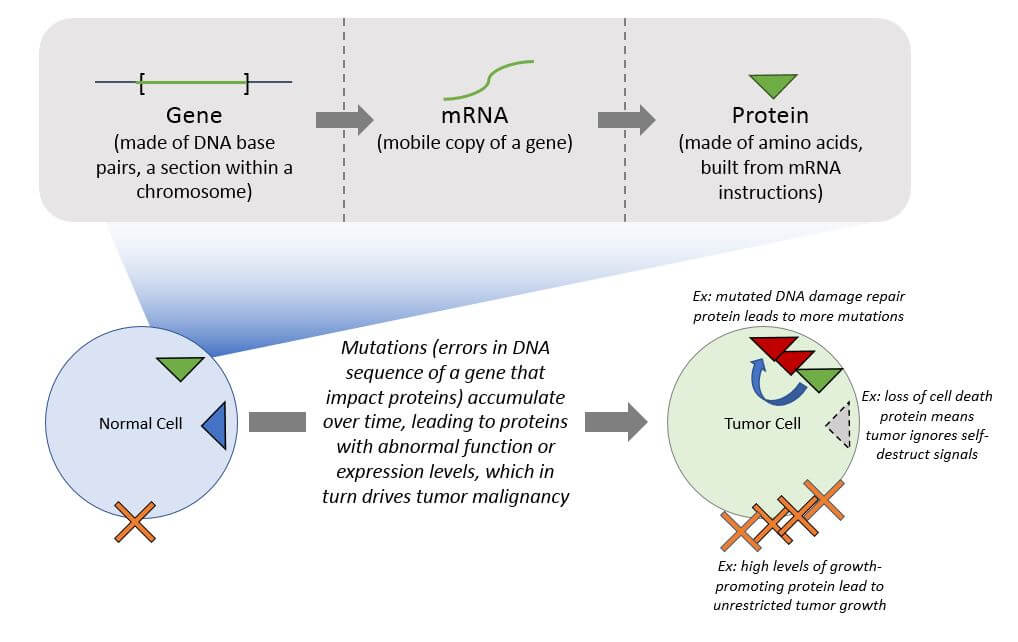
Cancer occurs when healthy cells experience a cascade of genetic errors (mutations) that shift normal biology in specific ways that drive malignancy. Examples include uncontrolled growth or the inability to repair DNA damage resulting in additional mutations. Disrupting these malignancy pathways without irrevocably damaging healthy tissues is the overarching therapeutic goal for all cancers. At its core, precision medicine oncology involves identifying and targeting cancer-specific molecular alterations.
Cancer is a particularly difficult disease to treat because each cancer arises in a unique fashion due to random mutations over time, and then tumor cells evolve to become resistant to whatever treatment is attempted. A treatment that is targeted to only part of a tumor, or to an older version of that tumor, will inevitably fail. This evolution leaves a “trail” of molecular alterations that tell the story of how the tumor formed, grew, and spread, and the core challenge of precision medicine is to efficiently and accurately measure these alterations in order to direct a more personalized treatment strategy.
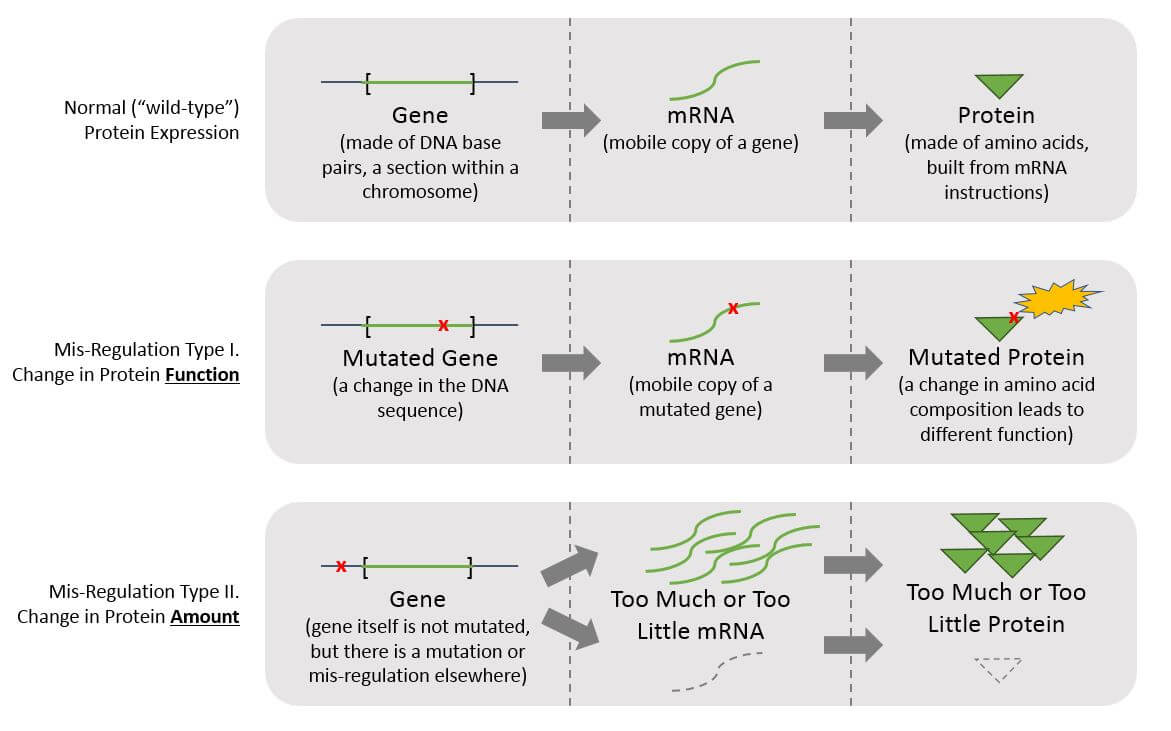
To accurately detect molecular alterations, three types of molecules must be considered: DNA, mRNA, and proteins. This requirement is rooted in the fundamentals of biology – the mis-regulations that drive cancer can be reflected at any one or more points in the following stepwise process of translating DNA sequence into mRNA into protein, so alterations in any or all three can contribute to a given tumor’s mis-regulation(s):
While we frequently refer to DNA or genes when discussing mutations, it is changes in protein function or dramatic differences in the amount of a non-mutated protein (either loss of expression, or over-expression) that are the molecular alterations ultimately responsible for cancer biology. We can therefore classify precision medicine tests based on which type of protein mis-regulation they can detect: A change in protein function, or a change in protein amount. Each of these can be detected at various levels (DNA, mRNA, and / or protein). To detect a previously unknown molecular alteration, multiple types of tests may be necessary.
Precision medicine test design therefore involves both a choice of molecule type (DNA, mRNA, protein) as well as what is being measured – an alteration in sequence (impacting protein function) or a change in expression levels (protein amount). The choice of molecule has implications on the capabilities of the testing technology.
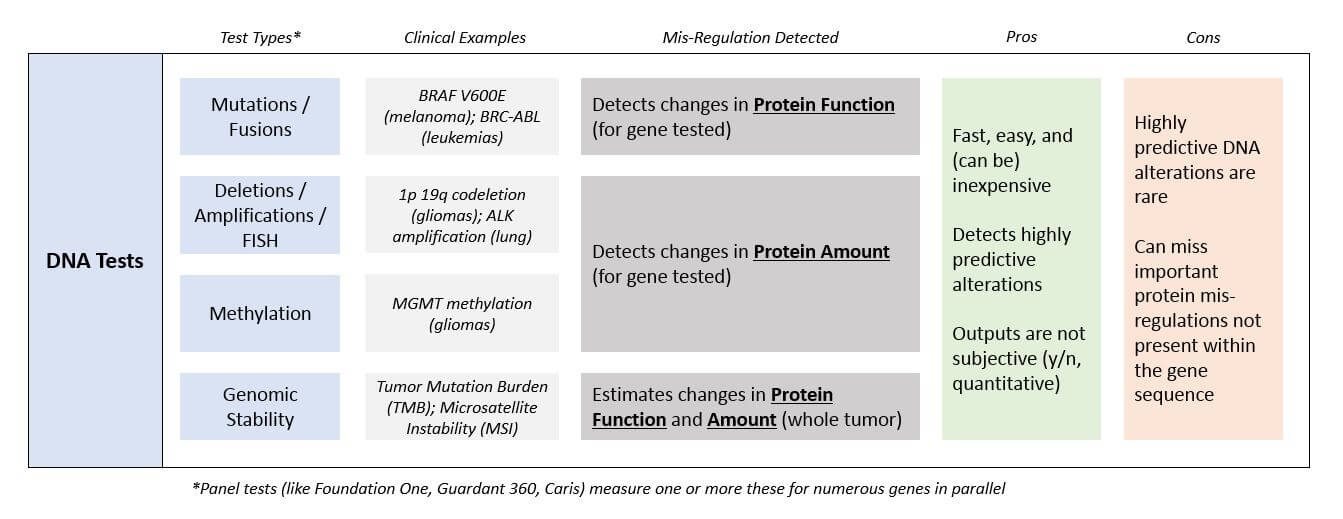
DNA Tests (Sequencing / Methylation / FISH)
These technologies detect and report out the presence or amount of a specific DNA sequence to identify any alterations within that gene. This includes changes to the sequence within a gene (mutations), as well as deletions, duplications, or fusions of portions of the gene. A DNA mutational test can be designed to test for one specific mutation that is common or suspected (like BRAF V600E testing in melanoma), or hundreds done in parallel to look more broadly for multiple mutations (like the pan-tumor Foundation One panel).
There is also a specific type of sequencing can also be done to detect DNA methylation, which is a chemical tag on the DNA itself near the gene that means the associated gene is “silenced” (won’t be expressed). This provides indirect evidence of changes in protein amount.
A Fluorescence In Situ Hybridization (FISH) test is when a fluorescent tag is used to mark specific sequences of DNA. It can detect deletions or amplifications, both of which predict changes in protein amounts.
Broad panels that search for numerous alterations can report out a list of all the mutations identified, as well as an overall count of how many alterations are seen in the entire genome. This is commonly referred to as genomic stability. Tumors that have numerous mutations are said to have a high tumor mutational burden (TMB), or to be microsatellite instable (MSI-hi). This measure predicts prognosis and also response to checkpoint inhibitor immunotherapies.
As compared to the other types of molecular tests, DNA tests require very little sample to test. They’re also fast and easy to interpret. If a test comes back positive for a known mutation, it is also likely to be clinically significant because an alteration within a gene will predictably impact the associated protein. However, a negative result does not mean that the associated protein is not involved in tumor formation or growth, because many times a mis-regulated protein does not have a mutation in its gene but is rather being impacted by another molecular alteration that is not necessarily detected by a given test.
DNA tests have been quite well-integrated into clinical practice in a few limited settings where certain mutations are common and where targeted drugs are available, for example non-small cell lung cancer and melanoma, where specific mutations (EGFR and ALK for lung; BRAF for melanoma) are near-universally tested for. DNA methylation tests are also used to indicate prognosis as well as sensitivity to certain treatments (MGMT methylation in brain tumors). The broader panels are also used in rare tumors or in other settings where the clinician or patient desires a more comprehensive search for mutations.
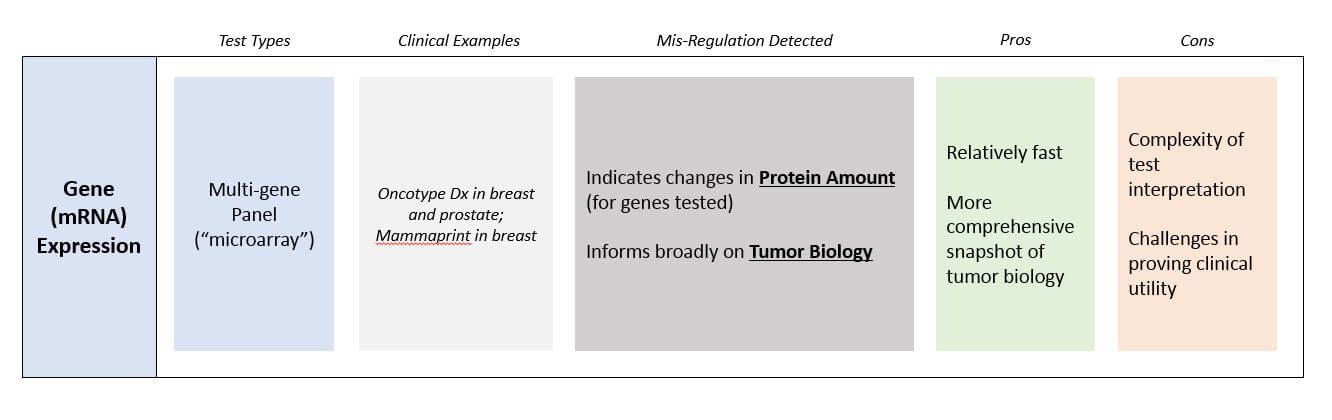
Gene Expression (mRNA) Testing
This technology measures the amount of mRNA of a gene. Like DNA sequencing, it is also relatively fast and inexpensive, although slightly more technically challenging than DNA sequencing. The interpretation is significantly more challenging because it is a relative and only semi-quantitative measure: It requires choosing a reference set of genes that is believed to be relatively stable to determine if the mRNA of interest is lower or higher than “normal”. Most gene expression tests therefore use a panel approach that tests dozens or hundreds of genes at once, so that any abnormal expression will be more obvious.
In terms of clinical utility, gene expression testing is typically bypassed in favor of DNA testing (which is easier) and protein testing (which more definitively reports out protein amounts). One notable exception is panel tests that help estimate the risk of recurrence for early stage breast and prostate tumors.
Gene expression tests do have enormous utility in research and drug development that may someday translate into broader clinical use, as they offer much more comprehensive snapshots of biology than either DNA or protein tests. In these settings gene expression tests are frequently used to better understand subtypes of a tumors and their underlying biology. This can lead to new targets for pharmaceutical development, as well as suggesting avenues for more personalized treatments.
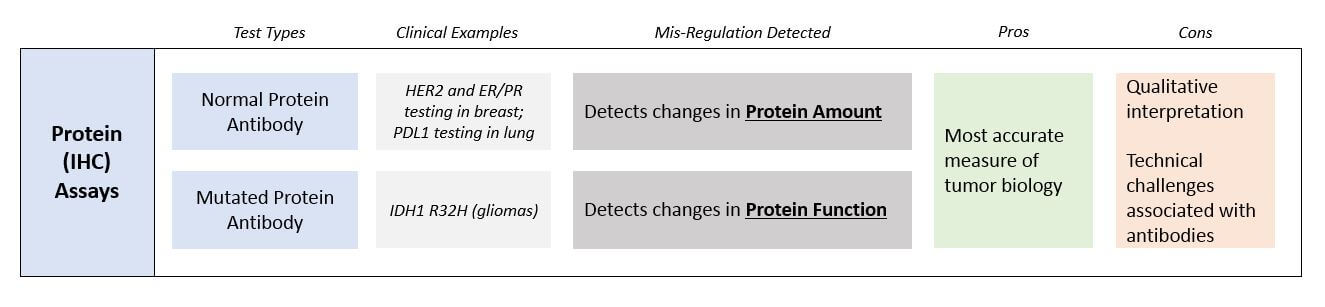
Protein Assays (IHC)
This technology, called immunohistochemistry or IHC, uses antibodies that bind to the protein of interest and can then be visualized or otherwise measured to provide a semi-quantitative measurement of target protein quantity. Antibodies can also detect specific protein mutations, such as an antibody to a mutated protein called IDH1 R132H is used in gliomas as a diagnostic and prognostic marker.
Protein assays are more technically challenging and expensive than either DNA or mRNA tests. This is because they are dependent on antibodies, which must be generated in cell culture or in live animals (and are harder to standardize and more likely to suffer from manufacturing issues). They also require significantly more sample to test. While DNA and mRNA can be extracted from minute, even single cell amounts, protein tests require much more tissue that must also be processed in a time-consuming way. Another challenge in protein assays is that because the antibodies are typically attached to fluorescent markers, the visible spectrum limits how many can be tested at once (most microscopes can only scan for a few distinct colors at once). Therefore, protein assays typically focus on just one or a few targets per test.
That said, because proteins are the effectors of biology, a direct measure of protein mis-regulation is very clinically relevant. Similar to DNA testing, there are several specific clinical settings where protein tests are already quite well integrated into clinical practice, most notably in breast cancer where ER, PR, and HER2 antibody testing is used to diagnose subtypes and direct targeted therapy treatment.
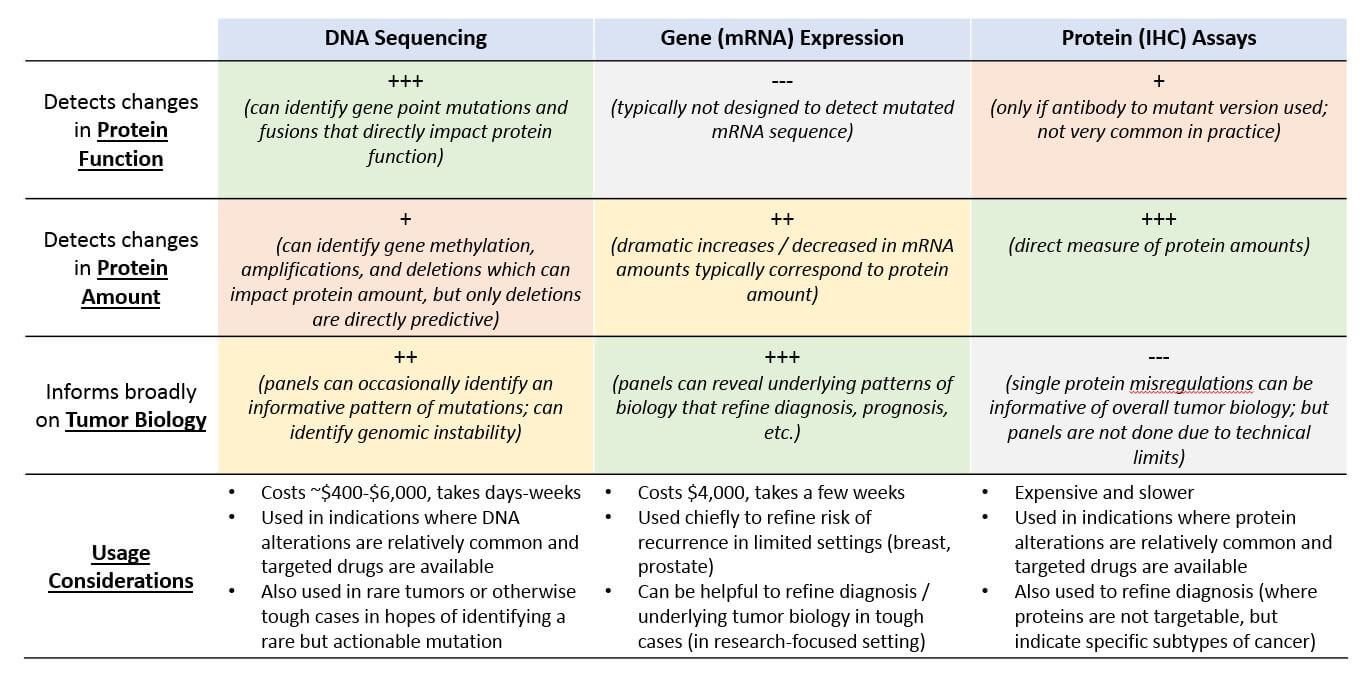
Molecular Alteration Tests Side-by-Side
Each type of testing technology measures different types of molecular alterations and provides its own snapshot of the underlying biology, and each has its pros and cons from both a scientific and a practical perspective. Roughly speaking, protein tests are the most biologically informative but also the most technically challenging and expensive. Single gene DNA-based tests cast a rather narrow net, as the known actionable mutations are few and far between. But, they’re much cheaper and provide a result faster. Typically, clinicians will start with a limited set of tests that are specific to the known biology of that tumor type. Then if they don’t detect anything, they may progress to broader panels in search of something rarer but still actionable.
Overview of Sample Sources and Implications
So, one key question is which type of molecular alteration to test for (DNA, mRNA, or protein). However, there is also the question of where and how to obtain the sample material to use for testing.
Sample source matters because
- Tumors can contain different cell populations in different spots
- The overall set of mutations and alterations within the tumor changes in response to treatment and over time during tumor growth
So, both the location of the sample as well as how recently it was obtained are critical considerations when interpreting the data.
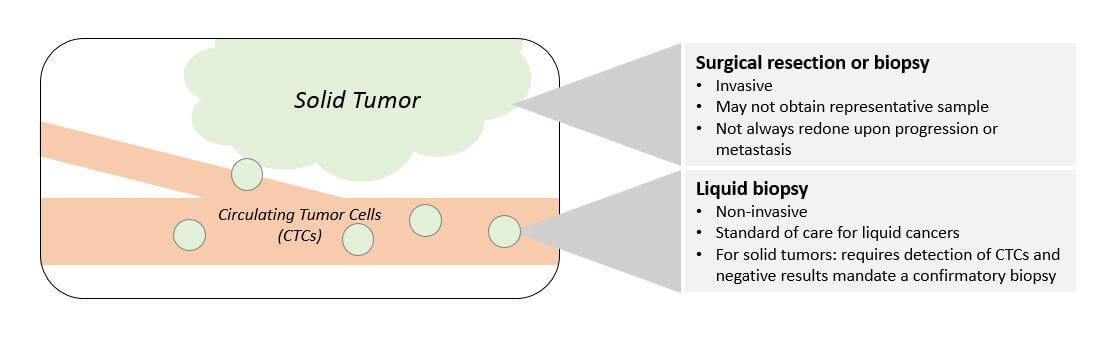
In the liquid cancers (leukemias and lymphomas), testing is relatively straightforward as the tumor cells are easily collected from the blood and can be tested frequently to track response and progression. Conversely, solid tumor testing has inherent challenges due to its invasive nature as well as the typically small amounts of tissue recovered, some of which must be reserved for pathology testing.
There is also a temporal issue: if the treatment works initially but the tumor ultimately becomes resistant and begins growing again or metastasizes, another biopsy is not always done, meaning there is no up-to-date sample for testing. Even if a biopsy is done, it is not always retested for molecular alterations. This gap in testing risks missing key information about resistance mechanisms that could be addressed with targeted approaches.
The emerging technology of testing Circulating Tumor Cells (CTCs), which are shed from solid tumors and end up circulating in the bloodstream, is an intriguing workaround to the issues surrounding solid tumor testing. This technology enables identifying and purifying CTCs from a typical blood draw (based on molecular tags seen on cancer cells but not healthy cells). Once purified, the CTCs are used for DNA-based tests. The Guardant 360 test which measures a broad panel of mutations, as well as more targeted tests like the Cobas T790M test, are already in clinical use for this purpose. However, not all tumors shed identifiable CTCs at the same rate, so a negative result in a CTC-based test requires a follow-up test to confirm using a standard biopsy.
Conclusion and Future Directions
Because molecular alterations arise in various and sometimes unpredictable ways during the course of tumor formation and growth, each of the various testing technologies has its own unique value in the clinical setting. In a world with limited resources (tissue for testing, as well as financial cost of the tests), the clinical challenge is deciding which test or which sequence of tests to pursue. In our next installation, we will delve into the inter-related challenges of how to design these tests to maximize clinical utility, and how clinicians currently navigate this complex landscape.